Research
My main research interest is to understand the evolutionary processes that produce adaptive phenotypic variation, particularly in situations where organisms are exposed to novel environmental conditions. I am tackling these questions both over short (phenotypic plasticity) and longer (adaptive evolution) time scales and my research is mostly focused on the gut microbiota associated with different lineages of fishes (i.e., threespine stickleback and Neotropical cichlids), but I occasionally explore other study systems (e.g., White Sands lizards).
The gut microbiota
We all contain multitudes: more than half of our body's cells are microorganisms. The recent realization that these microbes can play a crucial role for the ecology and evolution of their hosts is highly fascinating. The so-called gut microbiota is particularly complex and dynamic, and its composition is shaped and maintained by a combination of host genetics and environmental factors (e.g., diet). One of the main goals of my research is to understand how the gut microbiota changes during ecological divergence of host lineages. I am addressing this question in model systems in evolutionary biology such as threespine stickleback, Nicaraguan cichlids and White Sands lizards.
Quantifying gut microbiota (non)parallelism
|
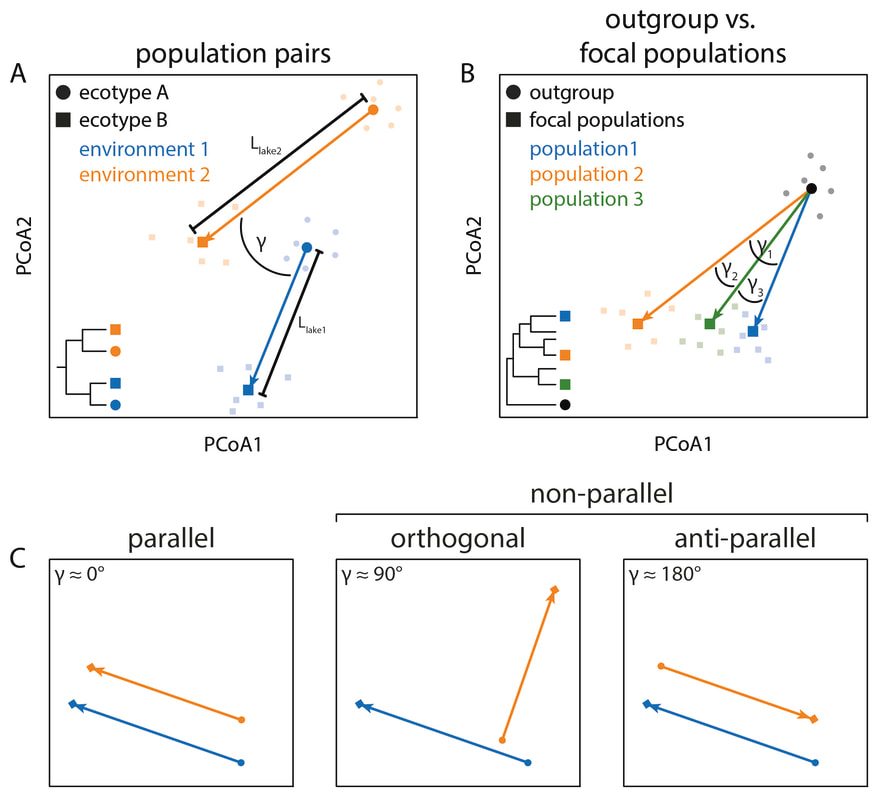
Parallel evolution of phenotypic traits is regarded as strong evidence for natural selection. Yet, we have limited knowledge of whether parallel evolution of host organisms is accompanied by parallel changes of their associated microbial communities which are crucial for host ecology and evolution. Determining the extent of microbiota parallelism in nature can improve our ability to identify the factors that are associated with (putatively adaptive) shifts in microbial communities. While it has been emphasized that (non)parallel evolution is better considered as a quantitative continuum rather than a binary phenomenon, quantitative approaches have rarely been used to study microbiota parallelism. Hence, we advocate using multivariate vector analysis to quantify direction and magnitude of microbiota changes (Ecology and Evolution 2022) and we compiled an R package for multivariate vector analysis of microbial communities (‘multivarvector’). This approach provides an analytical framework for quantitative comparisons across host lineages, thereby providing the potential to advance our capacity to predict microbiota changes. Hence, we emphasize that the development and application of quantitative measures, such as multivariate vector analysis, should be further explored in microbiota research in order to better understand the role of microbiota dynamics during their hosts’ adaptive evolution, particularly in settings of parallel evolution.
|
Multivariate vectors connect the population means (centroids) between host populations based on microbial community composition. Using these vectors, we can quantify the direction (γ) and magnitude (L) of microbiota changes associated with ecological and phylogenetic divergence of their hosts.
|
White Sands lizards of the Chihuahuan desert
|
|
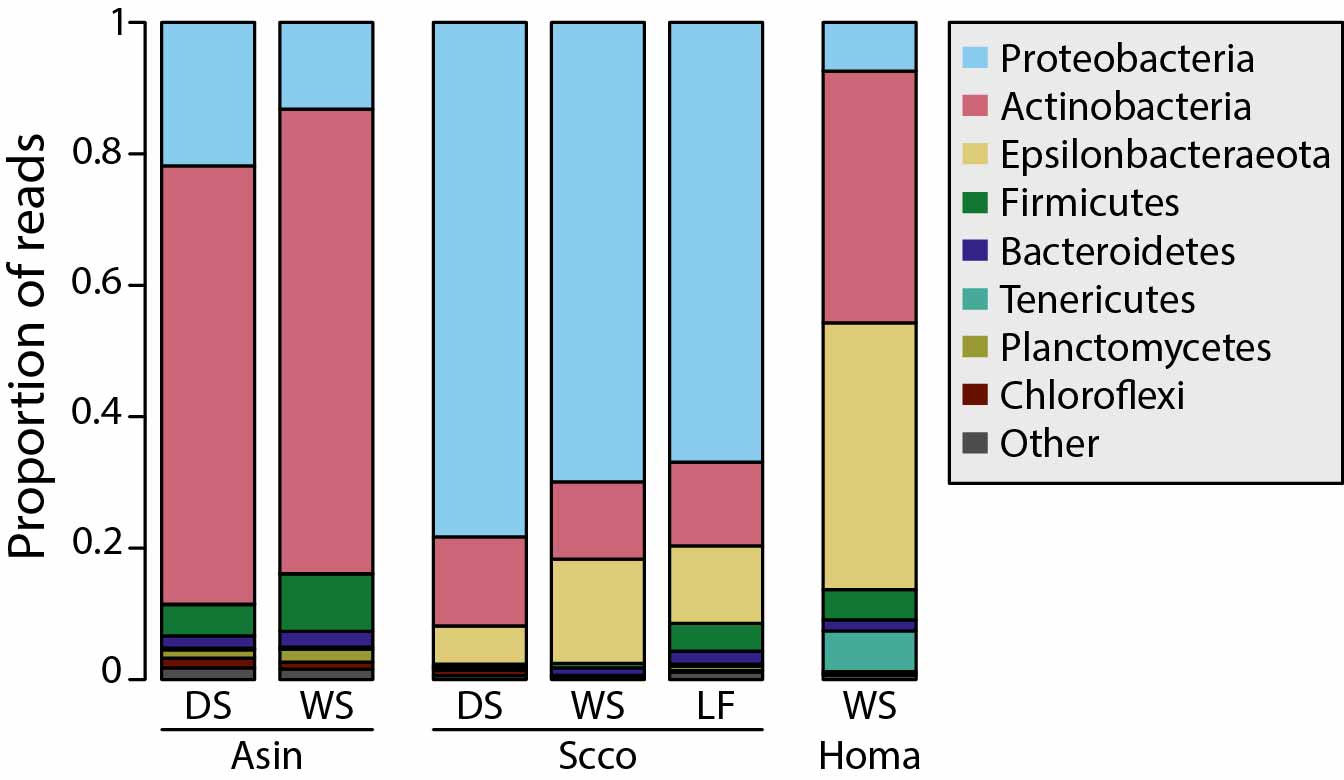
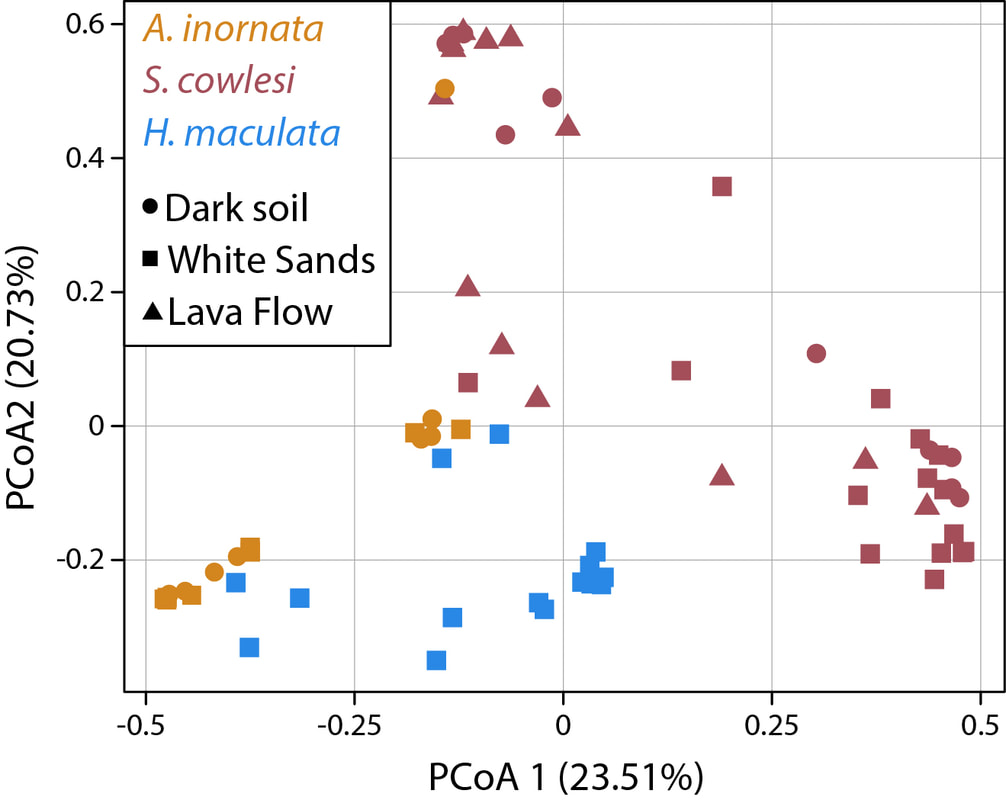
When organisms colonize novel environments, the newly encountered abiotic and biotic conditions impose selective pressures leading to ecological divergence as the colonizing populations adapt to the new environment. Natural settings in which independent lineages repeatedly colonized ecologically similar environments and evolved similar phenotypic traits (i.e., parallel evolution) can be useful models to investigate the contributions of deterministic and stochastic components of evolutionary change. We leveraged such a natural system in the Chihuahuan desert of New Mexico where three lizard species (Aspidoscleis inornata, Holbrookia maculata and Sceloporus cowlesi) colonized two geologically young and unique habitats: the White Sands and the Carrizozo lava flow (Mol Ecol 2023). We asked if and how the repeated colonization of novel habitats, and accompanying changes in trophic ecology, by independent host species affected their gut microbiota, and in particular, whether parallel host evolution led to parallel changes in gut microbiota composition. Host species generally had a stronger impact on gut microbiota composition than the lizards' environment and some gut microbiota changes were associated with shifts in diet after colonizing the White Sands habitat. Notably, colonization of the same environment by independent host species led to gut microbiota parallelism, whereas the colonization of two distinct environments by the same host species led to gut microbiota divergence. Such colonization events can improve our understanding of how host–microbiota interactions affect host ecology and evolution, potentially facilitating or constraining diversification.
|
Threespine stickleback fish from Vancouver Island, Canada
|
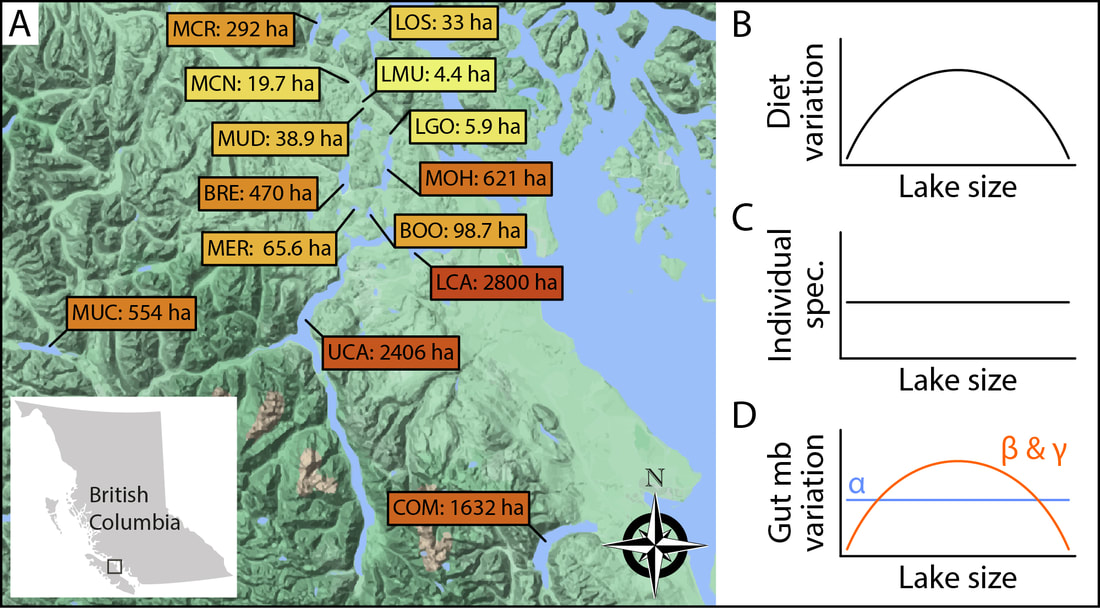
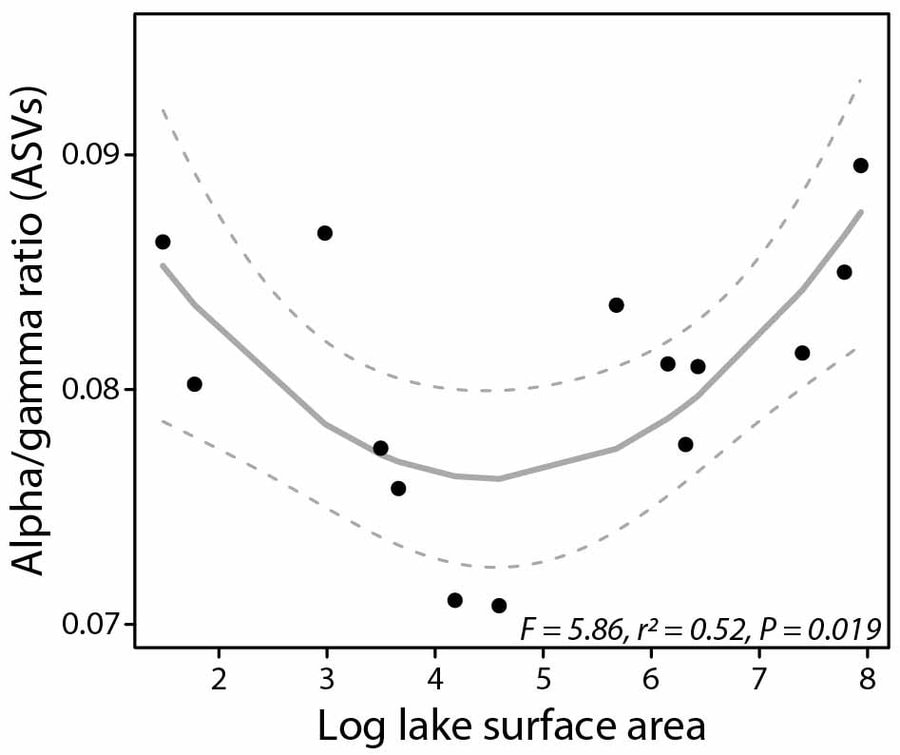
Threespine stickleback (Gasterosteues aculeatus) are widespread across the Northern hemisphere and are considered a textbook example of parallel evolution associated with the repeated colonization of and adaptation to freshwater habitats. On Vancouver Island, Canada, stickleback show substantial variation in their trophic ecology across a large number of lakes, which makes them a powerful system to study gut microbiota dynamics in response to novel environmental conditions (e.g., different diets). My research is focused on the question of how the interaction between host and gut microbiota can facilitate the exploitation of novel trophic niches. To tackle this question in a comprehensive manner, I am studying gut microbiota variation within and among wild populations as well as performing controlled experiments in the lab. In a first study of wild populations from 14 lakes, we tested whether trophic niche width is associated with gut microbiota diversity, divergence and uniqueness on the host population level (Am Nat 2023). For this, we used lake size as a proxy for trophic ecology because the proportions of benthic and limnetic diet have been shown to vary with lake size in stickleback. We found higher gut microbiota uniqueness among individuals from populations with broader trophic niches, suggesting that diet diversity promotes gut microbiota diversity. These exciting results provide a starting point for the further exploration of how diversity of resources, hosts and their gut microbiota might affect host adaptation. Addressing this question can improve our understanding of the eco-evo consequences of host-microbiota interactions.
|
|
The young adaptive radiation of Nicaraguan cichlid fish
|
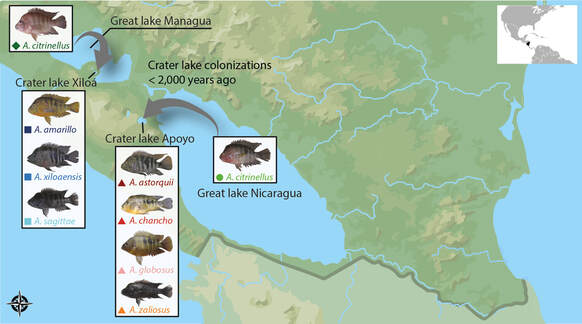
To understand how the gut microbiota changes during early stages of ecological divergence and speciation, I investigated the gut microbiota and trophic ecology of multiple species from two young adaptive radiations of Nicaraguan cichlids. We found that differentiation in diet is associated with gut microbiota differentiation, suggesting that diet affects the gut microbiota (Microbiome 2020). Yet, across two crater lakes, we did not find strong evidence for parallel gut microbiota changes. This might be explained by a lack of sufficient trophic niche differentiation among these young species.
|
The visual system
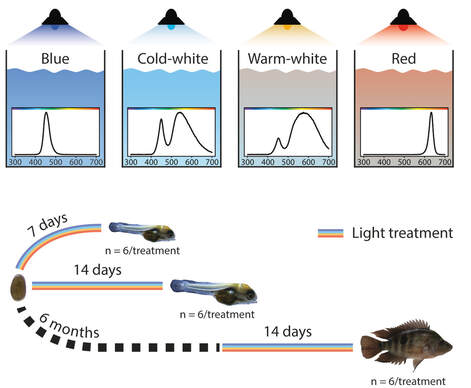
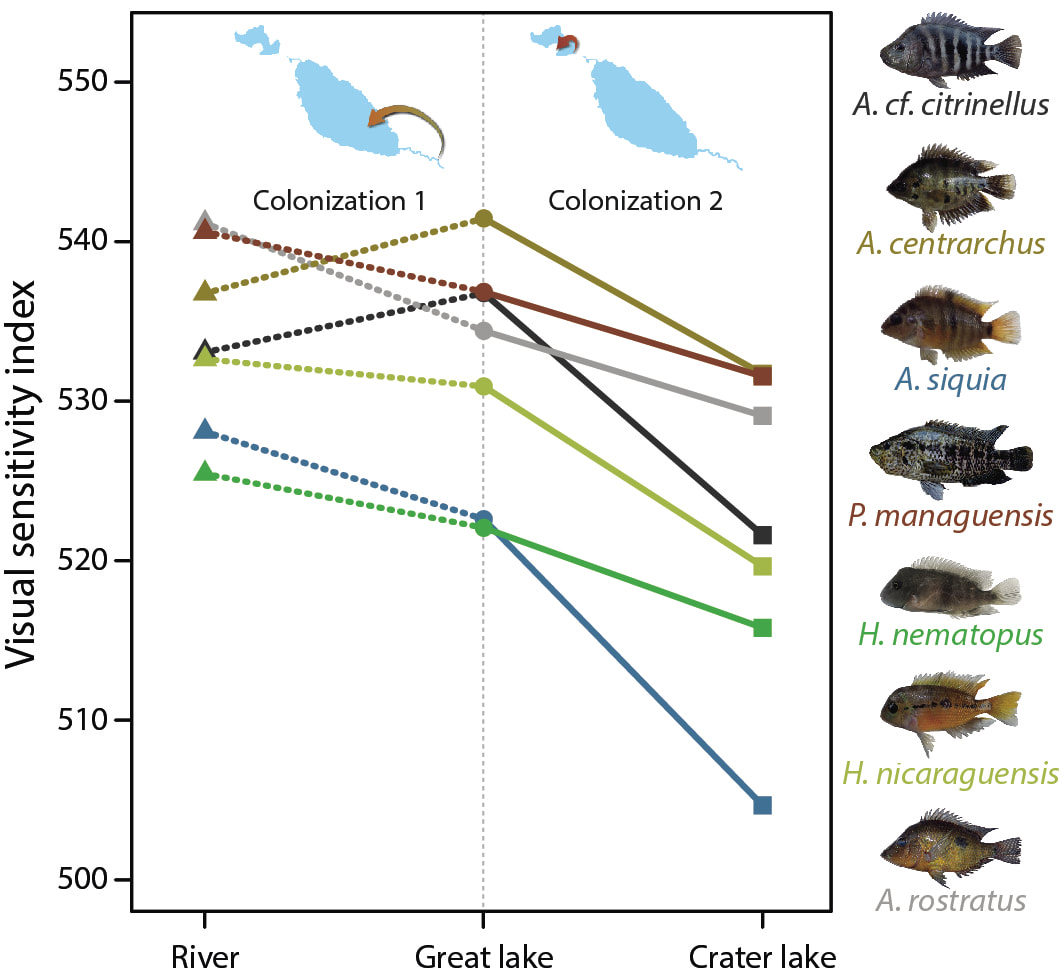
My research on the visual system aims at understanding how changes on the molecular level are translated into adaptive phenotypic variation. The visual system of cichlid fish represents an excellent model to study adaptive evolution since it is highly variable, the phenotypic effects of molecular changes are well-understood and there is detailed knowledge on the adaptive value of visual phenotypes under certain environmental conditions. Our work on the visual system has demonstrated that, at least over short evolutionary scales, regulatory changes are more important than structural changes during adaptation to novel light conditions. At this time scale, the direction of phenotypic change is largely predictable, although the particular molecular mechanisms underlying such changes might not be (MBE 2017, Evol Let 2018). Notably, shifts in gene expression during ontogeny appear to play a crucial role during adaptive evolution of the visual system of cichlids (Mol Ecol 2017, RSOS 2019). Moreover, the ability to adjust the visual system to changing light conditions can occur very rapidly and appears to be maintained into adulthood (JEZ-B 2018). Overall, this work has improved our knowledge on how cichlids can adjust their visual system to novel light conditions almost instantaneously via phenotypic plasticity but also very rapidly (on an evolutionary time scale) over a few hundred generations via adaptive evolution.
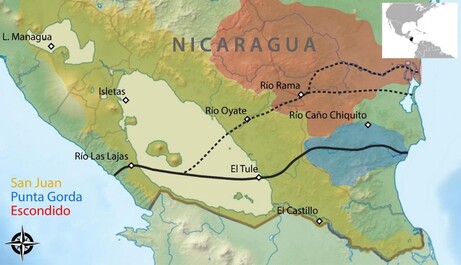
The Nicaragua Canal
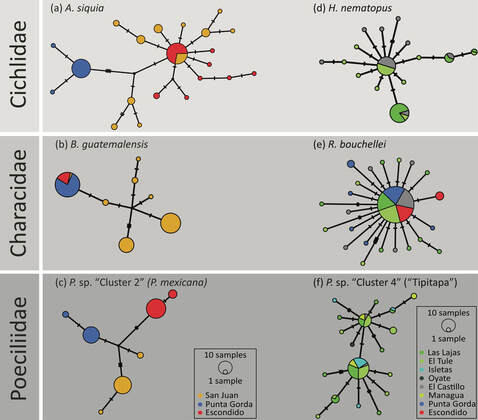
In 2013, a Chinese consortium was granted the concession to build an interoceanic shipping canal, called the Nicaragua Canal, across the Central American isthmus through Nicaragua. Such large-scale infrastructure projects commonly have drastic effects on ecosystems, thereby threatening the maintenance of biodiversity. We sought to shed light on species’ distribution and patterns of population structure across distinct watersheds that would be connected by the canal and intended to inform about the effects of artificially connecting these watersheds on the freshwater fish fauna of Nicaragua (Conserv Biol 2017). We found that only one third of freshwater fish species are shared among the two watersheds whereas one third exclusively occurs in either of the two. This emphasizes on the huge threat of biotic homogenization once the two watersheds would be connected. Further, mitochondrial DNA (cytb) sequencing of multiple species revealed that populations from distinct watersheds are genetically differentiated whereas within the same watershed, genetic differentiation is commonly low. Bringing these differentiated and locally adapted populations into contact through the Nicaragua Canal would most likely cause an extensive loss of genetic diversity. Further, species were detected in areas where they were not known to exist. Taken together, these results demonstrate that constructing this canal would have adverse effects on Nicaragua’s freshwater biodiversity. Fortunately, the construction of the Nicaragua Canal has not started yet and, most likely, this risky and irrational project won't be realized in the near future.